Background & Aim
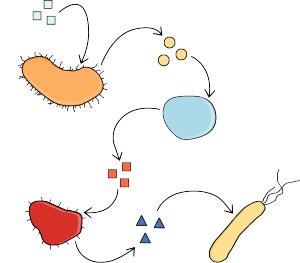
In the human gut lives a myriad of bacterial species, known as the gut microbiome, which is of immense importance for health and development [1, 2]. Changes in its composition have been linked to diseases such as diabetes [3] and inflammatory bowel disease [4], suggesting that manipulation of the gut microbiome might have potential in disease prevention and treatment. To make use of this potential, we must increase our understanding of the processes that influence microbiome assembly and functioning.
The gut microbiome is largely characterized by cross-feeding interactions, where microbes consume metabolites produced by another individual [5]. These interactions often occur in multiple consecutive steps, such that organism A creates a byproduct consumed by organism B, which in turn creates a byproduct consumed by organism C, and so on. A recent study has found that the average length of such cross-feeding chains in the human gut are four steps [6]. However, mappings of metabolic capabilities of present species reveal that much longer chains are possible. Thus, it is not yet known why these chains are so severely constrained, when they need not be in principle.
Aim
In this study, I aim to investigate if recurring perturbations constrain the length of cross-feeding chains. To answer this question in as much generality as possible, I use a system of digital evolution, where self-replicating computer programs evolve by means of natural selection.
Continue to Methods.
References
- Cho, I., Blaser, M. J., 2012. The human microbiome: at the interface of health and disease. Nature Reviews Genetics 13, 260–270.
- Brestoff, J. R., Artis, D., 2013. Commensal bacteria at the interface of host metabolism and the immune system. Nature Immunology 14, 676–684.
- Qin, J., et al., 2012. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60.
- Franzosa, E. A., et al., 2019. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nature Microbiology 4, 293–305
- Sung, J., et al., 2017. Global metabolic interaction network of the human gut microbiota for context-specific community-scale analysis. Nature Communications 8, 15393.
- Wang, T., Goyal, A., Dubinkina, V., Maslov, S., 2019. Evidence for a multi-level trophic organization of the human gut microbiome. PLOS Computational Biology 15, 1–20.
Responsible for this page:
Director of undergraduate studies Biology
Last updated:
05/28/21
